Quantitative Structure Property Relationships (QSPR)
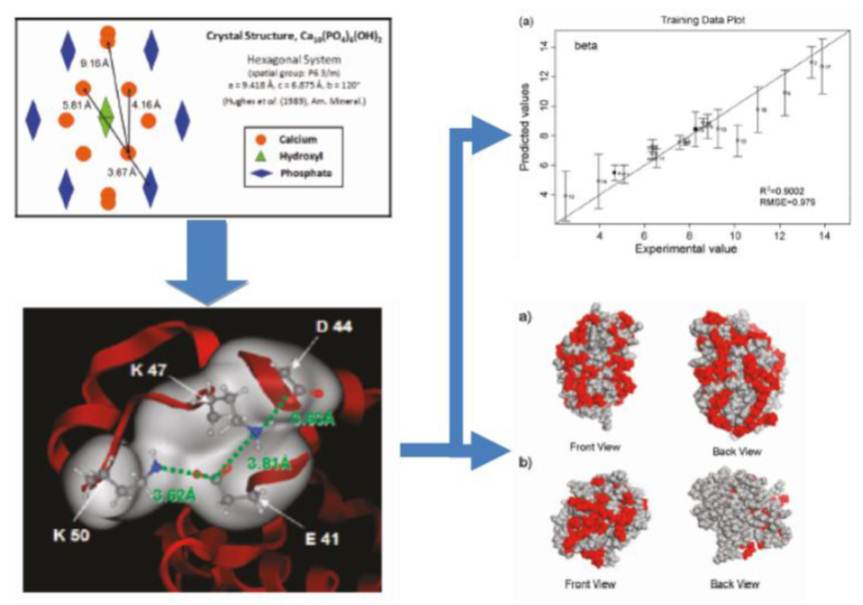
QSPR is powerful analytical method for breaking down a molecule into a series of numerical values describing its relevant chemical and physical properties (e.g. charge density, hydrophobic surface area, etc.). Using fundamental insights about protein-ligand interactions (i.e. the spacing of functional groups in the crystal structure of ceramic hydroxyapatite), we can also create specialized numerical descriptors that calculate the favorable resin binding sites on the protein and then use them as input values for a predictive model or for protein surface visualization.
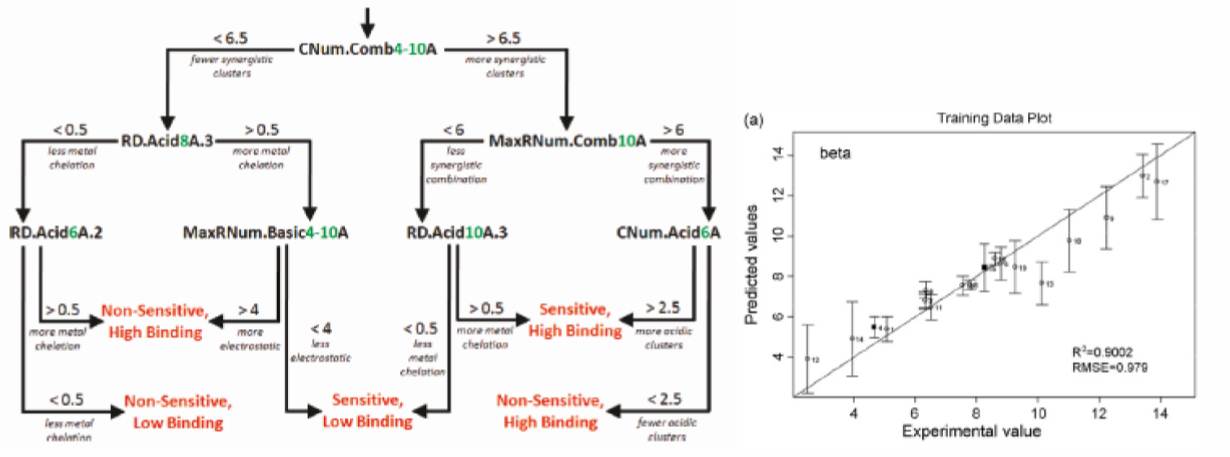
In order to convert these numerical descriptors for ligands and/or their protein targets into a protein retention model, we use machine-learning methods that first select relevant variables from the pool of descriptors and then used either linear or non-linear operations to combine selected descriptors into a predictive model. These models can either predict the class of retention behavior expected for each protein (shown left) or a numerical value for the protein’s retention in a particular chromatographic system (shown right).
Previously, we have developed models for a variety of chromatography systems (IEX, HIC and hydroxyapatite) and are currently developing a model for multimodal systems. To this end, we are designing set of novel descriptors, based on the insights gained from MD simulations, to identify relevant protein surface sites for binding multimodal ligands. Using our current libraries of descriptors, we are also generating machine-learning models to predict the binding affinity for novel affinity peptide systems.
Selected References:
- Y. Hou, C.J. Morrison and S.M. Cramer. Anal. Chem. (2011). Classification of Protein Binding in Hydroxyapatite Chromatography: Synergistic Interactions on the Molecular Scale.
- Y. Hou and S.M. Cramer. J. Chrom. A (2010). Evaluation of selectivity in multimodal anion exchange systems: a-priori prediction of protein retention and examination of mobile phase modifier effects.
- Y. Hou, et al. Biotechnol. Bioeng. (2010). Improved Process Analytical Technology for Protein A Chromatography Using Predictive Principal Component Analysis Tools.
- J. Chen, T. Yang and S.M. Cramer. J. Chrom. A (2008). Prediction of protein retention times in gradient hydrophobic interaction chromatographic systems.
