Nuclear Magnetic Resonance (NMR)
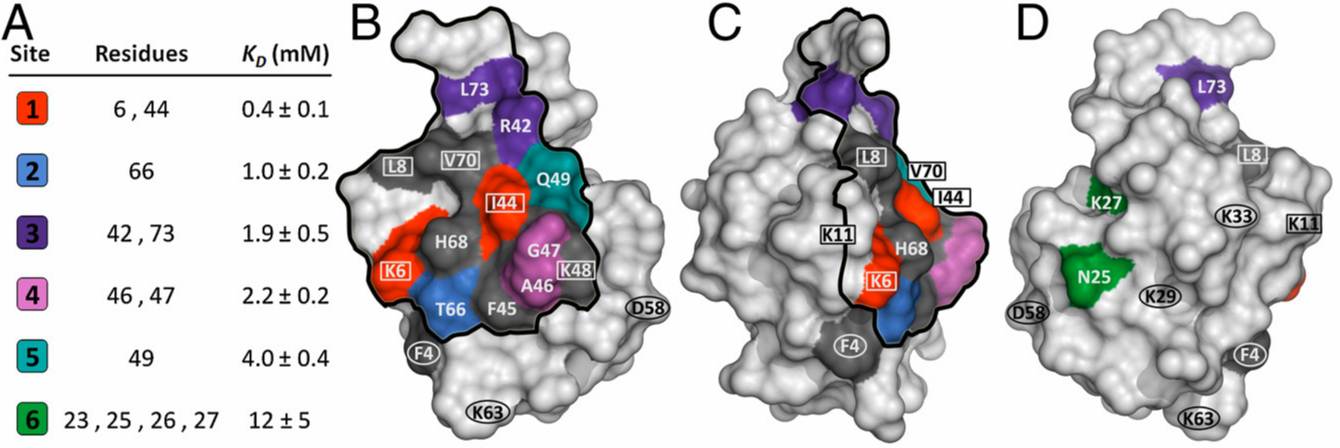
Nuclear magnetic resonance is widely employed for protein structure determination and measuring protein-target interactions. In our studies, two-dimensional heteronuclear single quantum correlation (HSQC) experiments are employed to study the interactions of proteins with ion exchange and multimodal chromatographic ligands in solution. This technique makes it possible to experimentally confirm the ligand interaction sites on a protein surface and the apparent binding affinities for each amino acid residue.
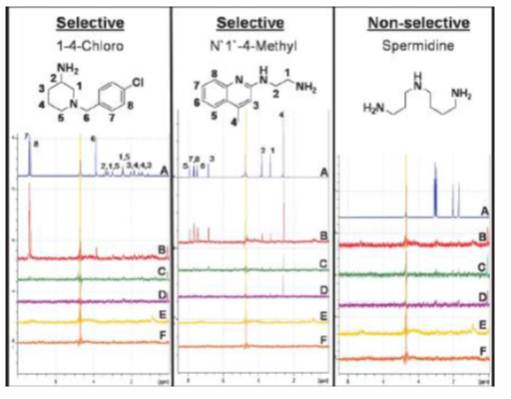
Saturation transfer difference (STD) NMR experiments are routinely used for examining protein-ligand binding in solution and this technique is often used as an initial screening technique for evaluating protein-ligand interactions and identifying the chemical moieties involved in binding. Both HSQC and STD-NMR can also be performed in the presence of fluid phase modifiers and/or displacers to probe the effect of these additives on the binding strength of different ligand moieties and elucidate a mechanism of action for these molecules.
Selected References:
- M.A. Holstein, et al., J. Chromatogr. A (2012). Probing multimodal ligand binding regions on ubiquitin using nuclear magnetic resonance, chromatography, and molecular dynamics simulations.
- W.K. Chung, et al. PNAS (2010). Evaluation of protein adsorption and preferred binding regions in multimodal chromatography using NMR.
- C.J. Morrison, et al. Biotechnol. Bioeng. (2009). Mechanistic Studies of Displacer-Protein Binding in Chemically Selective Displacement Systems Using NMR and MD Simulations.
