Purification of Proteins using Ion Exchange Chromatography - Senior Lab (Spring Semester)
Course Outline:
Goal: The objectives of this lab are:
- experimentally determine the adsorption isotherm parameters for the three proteins used in this lab;
- perform linear gradient column simulations outside of the lab to establish conditions that are amenable to separating the proteins of interest using the determined isotherm parameters and satisfying specified constraints; and
- validate these simulation results using a linear gradient experiment.
Introduction: In this senior laboratory you will be performing experiments and simulations of the chromatographic separations of proteins, similar to those performed in the biotechnology industry. Chromatography is a separations technique based on differences in the adsorptive or partitioning properties of mobile phase feed components with a porous stationary phase chromatographic material. The migration rate of the solutes is determined by the equilibrium distribution of components between the flowing fluid and the stationary porous medium. In this lab, you will be using a mode of chromatography called ion exchange chromatography which separates proteins based upon their electrostatic interactions with a charged chromatographic material (Cramer and Natarajan, 1999). In the first part of the lab, isocratic (constant salt concentration) experiments will be performed on a BioRad chromatographic workstation to determine the adsorption isotherm parameters for the proteins. After experimentally determining these parameters, you will employ the parameters (outside of the lab) in a chromatographic simulation software to determine an optimal linear gradient separation method. In the final part of the lab, the students will return to the lab to experimentally validate these simulated separation conditions, again using the BioRad chromatographic workstation. The protein system for these separations will consist of three commercially available proteins (α-chymotrypsinogen A, ribonuclease A, and lysozyme). The column to be used for the separation contains sulfopropyl sepharaosefast flow (SP Sepharose FF, GE Healthcare). For the simulations, the Steric Mass Action (SMA) isotherm for protein ion exchange chromatography will be employed. This isotherm was developed by Brooks and Cramer (1992) and enables the prediction of multicomponent protein equilibrium at various salt concentrations. This isotherm has been shown to accurately describe the behavior of proteins in ion exchange systems under isocratic elution (Gallant et al 1995), linear gradient (Gallant et al 1996), step gradient (Gallant et al 1995) and displacement (Natarajan et al 2000) chromatographic modes of operation.
The single component SMA isotherm is given by:
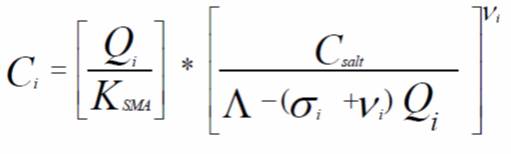
Where Ci is the concentration of species in the mobile phase, Qi is the concentration on the stationary phase, KSMA is the SMA equilibrium constant of the ith component, Csalt is the concentration of the counter ion, Λ is the total bed ionic capacity of the stationary phase, σi is the steric factor, and vi is the characteristic charge.
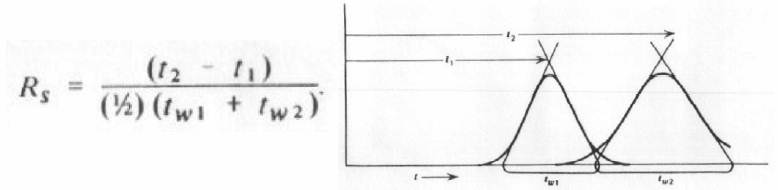
There are three isotherm parameters for each protein: KSMA, vi, and σi. The KSMA and vi parameters are considered “linear” isotherm parameters since they can be determined directly from experiments carried out in the linear region of the isotherm (i.e. at low mobile phase concentrations). These are the two parameters that will be experimentally determined for each protein in this laboratory. The steric factor σi is the nonlinear isotherm parameter and describes the behavior at high solute concentrations. The following nonlinear SMA parameters (which due to time constraints and material costs will not be measured experimentally) can be assumed for this laboratory: σ(α-chymotrypsinogen A) = 31.7, σ(ribonuclease A) = 17.2, and σ(lysozyme) = 17.0. The liquid chromatography column packed with the SP Sepharose FF stationary phase has a length of 10 cm and a column inner diameter of 0.46 cm. Further, the resins can be assumed to have the following properties: interstitial porosity (εe) of 0.40, intraparticular porosity (εp) of 0.70, bed capacity (Λ) of 525mM. A useful concept that you will need to use in this lab is the resolution of two peaks. The equation for the resolution between two chromatographic peaks (Rs) with retention times of t1 and t2, respectively is given below where tw1 and tw2 represent the widths at the bases of the two peaks (note: these widths can be determined either directly from the baseline or by drawing lines through the inflection points as indicated in the figure). The figure at the right below gives a graphical representation of what the terms in the equation relate to.
The total porosity (εT) of a chromatographic column can be calculated using the equation below where the term (εp) represents the intraparticular porosity and (εe) represents the interstitial porosity of the column.
εT = εe + (εp)( 1 - εe )
The chromatographic efficiency of the column can be calculated using any of the peaks from the chromatogram. The efficiency of the column is related to the number of theoretical plates in the column, N, which is equivalent to 16(tr/tw)2 where tr equals the retention time of the peak and tw is equivalent to the width of the base of this peak, in units of time. Alternatively, N can be obtained as 5.54 (tr/tH)2 where tH is the width of the peak at half of the maximum peak height.
Expectations
Interim Discussion. Please come prepared to discuss your isocratic chromatographic data as well as the linear gradient simulations results using the chromatographic software. Please bring your experimental chromatograms, efficiency calculations, resolution calculations, retention results for an unretained solute, log (k’) plot, isotherms at each salt concentration, and simulated chromatogram. Be prepared to discuss the key concepts related to this laboratory. At our Interim Discussion session, we will discuss report details further.
Literature Cited
Brooks, C. and Cramer, S.M., Steric mass action ion exchange displacement profiles and Induced salt gradients, AIChE Journal 38 (12), 1969-1978, 1992.
Gallant, S.; Kundu, A. and Cramer, S.M.,Modeling nonlinear elution of proteins inion-exchange chromatography, Journal of Chromatography, 702, 125-142, 1995.
Gallant, S.; Kundu, A. and Cramer, S.M., Optimization of step gradient separations - consideration of nonlinear adsorption, Biotechnology and Bioengineering, 47, 355-372, 1995.
Gallant, S.; Vunnum, S. and Cramer, S.M., Optimization of preparative ion-exchange chromatography of proteins-linear gradient separations, Journal of Chromatography, 725,295-314, 1996.
Cramer, S.M. and Natarajan, V., Chromatography, ion exchange, in the Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation, 612-627, 1999.
Natarajan, V. and Cramer, S.M., A methodology for the characterization of ion-exchange resins, Separ. Sci. Technol., 35 (11) 1719-1742, 2000.
Natarajan, V.; Bequette, B.W. and Cramer, S.M., Optimization of ion exchange displacement separations. I. Validation of an iterative scheme and its use as a methods development tool, J. Chromatogr. A, 876 (1-2) 51-62 April 21, 2000.
